Advanced recipes
advanced_recipes.RmdThis vignette gives some examples of more advanced/involved uses of STbayes.
Dynamic network models
Dynamic networks that change over the observation period may also be
input by adding an optional column time containing integers
indicating which time-step the edges belong to. The network should be
summarized for each inter-event period. For example, if the user’s
observation period included 10 events and the dataset does not contain
censored individuals, they should supply edge weights from 10 networks
in total, where time=1 should contain the network
representing the period from
,
time=2 represents
,
and time=10 represents from
.
if the user’s observation period included 10 events and the dataset does
contain censored individuals, they should supply edge weights from 11
networks in total, where time=1 should contain the network
representing the period from
,
time=2 represents
,
and time=11 represents from
.
NB: If there are censored individuals, the end of the observation period
should necessarily be larger than the time of the final event
(event_data$t_end > max(event_data$time).
In the toy example below, we create a dynamic foveation network (indicating whether or not the focal individual foveated at the other individual) in a group of 3 individuals. None of the individuals are censored, so we only need to provide networks summarized over three timesteps:
networks <- data.frame(
trial = 1,
focal = c("A", "A", "B",
"A", "A", "B",
"A", "A", "B"),
other = c("B", "C", "C",
"B", "C", "C",
"B", "C", "C"),
foveation = c(1, 0, 1, #summarizes from start of obs period to first event
0, 1, 1, #summarizes from first event to second event
0, 0, 1), #summarizes from second event to final event,
time = rep(1:3, each=3)
)Multi-network models
To create a multi-network model, you can just add more columns to the
networks data-frame. Below is a toy data-set of two trials
each with 3 individuals, and we define a kin and inverse distance
network.
event_data <- data.frame(
trial = rep(1:2, each = 3),
id = LETTERS[1:6],
time = c(2, 1, 3, 4, 2, 1),
t_end = c(3, 3, 3, 4, 4, 4)
)
networks <- data.frame(
trial = rep(1:2, each = 3),
focal = c("A", "A", "B", "D", "D", "E"),
other = c("B", "C", "C", "E", "F", "F"),
kin = c(1, 0, 1, 0, 1, 1), # first network
inverse_distance = c(0, 1, .5, .25, .1, 0) #second network
)
data_list <- import_user_STb(
event_data = event_data,
networks = networks)When fitting a multi-network model, it will estimate an value for each network. If you are using a complex transmission function, it will also estimate separate or parameters for each network.
Individual-level variables (ILVs)
Individual-level variables can be either constant (e.g. age, sex) or time-varying (e.g. body condition). Below I define constant ILVs age, sex and weight to add to our toy dataset above.
ILV_c <- data.frame(
id = LETTERS[1:6],
age = c(-1, -2, 0, 1, 2, 3), # continuous variables should be normalized
sex = as.factor(c("F", "M", "M", "M", "F", "F")), # Categorical ILVs must be input as factors
weight = c(0.5, .25, .3, 0, -.2, -.4)
)The data-frame ILV_c holds the values for each individual. The
formatting of id should match the event_data and networks
data-frames. At the moment, ILVs can be continuous, boolean or
categorical. Continuous variables should be normalized, and categorical
variables should be input as factors.
We need to explicitly tell STbayes which variables are additive (acting independently on intrinsic or social rates) and multiplicative (same effect estimated for intrinsic and social rates). Below, I have specified age as acting independently on the intrinsic and social rate, sex as acting only on the social rate, and weight as a multiplicative effect. Two betas will be estimated for age, and a single beta will be estimated for sex and weight.
data_list <- import_user_STb(
event_data = event_data,
networks = networks,
ILV_c = ILV_c,
ILVi = c("age"),
ILVs = c( "age", "sex"),
ILVm = c("weight")
)Time-varying ILVs
Below we add distance from resource as a time-varying ILV. Similar to
dynamic networks, these need to be summarized per inter-event interval.
For example, the value of dist_from_resource at
time=1 should reflect the average distance of the
individual to the resource from the start of the observation period to
the first event.
ILV_tv <- data.frame(
trial = c(rep(1, each = 9),rep(2, each = 9)),
id = c(rep(LETTERS[1:3], each=3), rep(LETTERS[4:6], each=3)),
time = c(rep(1:3, times = 3), rep(1:3, times=3)),
dist_from_resource = rnorm(18)
)
data_list <- import_user_STb(
event_data = event_data,
networks = networks,
ILV_c = ILV_c,
ILV_tv = ILV_tv,
ILVi = c("age", "dist_from_resource"),
ILVs = c("sex"),
ILVm = c("weight")
)Here, we will include it as an additive effect on the intrinsic rate along with age.
Transmission weights
Transmission weights are vital information that can dramatically improve how the model fits the data. NBDA models have supported static transmission weights (), which are usually some rate of behavioural usage. In the context of cultural transmission, individuals who rarely use a novel behaviour have a much lower chance of serving as demonstrators for others compared with individuals who frequently use it. In a study of disease transmission, this might be something like shedding rate. Transmission weights must be 0 or positive: , where we divide the number of behaviours produced by how the duration that individual is knowledgeable. We just need to define a dataframe with this information. For static transmission weights, we define:
t_weights_static <- data.frame(
id = LETTERS[1:6],
t_weight = rbeta(6, 1, 5)
)STbayes introduces the possibility of using dynamic transmission
weights, which can change over time. Dynamic transmission weights
are defined with an additional time column. Similar to
dynamic networks, these integer times correspond to the inter-event
intervals.
are the number of behaviours produced during interval / duration of
interval:
t_weights_dynamic <- data.frame(
trial = c(rep(1, each = 9),rep(2, each = 9)),
id = c(rep(LETTERS[1:3], each=3), rep(LETTERS[4:6], each=3)),
time = c(rep(1:3, times = 3), rep(1:3, times=3)),
t_weight = rbeta(18, 1, 5)
)These can be included in the model by:
data_list <- import_user_STb(
event_data = event_data,
networks = networks,
t_weights = t_weights_dynamic
)You can mix and match static/dynamic networks with static/dynamic transmission weights. Dynamic transmission weights are multiplied with the network edges to modulate their effect. Users should only include them if they are certain that they reflect real production rates, since a 0 value will remove any social influence that individual has on others for that timestep. This feature is designed for studies with automated methods that allow for accurate counts of behavioural productions. With coarser sampling regimes, users should instead use static transmission rates.
Varying effects by individual and trial
You may estimate varying effects on the intrinsic rate (lambda_0),
social learning rate (s) and any of the other parameters or ILVs.
Specify specific parameter names using the argument
veff_params in the call to
generate_STb_model(), and specify whether you want by
individual, trial or both using the veff_type argument:
model = generate_STb_model(data_list,
veff_params = c("lambda_0", "s"),
veff_type="id") #on id
model = generate_STb_model(data_list,
veff_params = c("lambda_0", "s"),
veff_type="trial") #on trial
model = generate_STb_model(data_list,
veff_params = c("lambda_0", "s"),
veff_type=c("id", "trial")) #on bothFor
,
and
,
varying effects are added onto the main effect prior to transformation
from log-scale back to linear. For example, if we apply a varying effect
for
,
the model will calculate a value for each individual in the
transformed parameters block of the Stan model:
and use those values when calculating
the likelihood in the model block. This ensures that
or
values are constrained to be positive.
You can also apply varying effects to ILVs, if you want:
event_data <- data.frame(
trial = rep(1:2, each = 3),
id = LETTERS[1:6],
time = c(2, 1, 3, 4, 2, 1),
t_end = c(3, 3, 3, 4, 4, 4)
)
networks <- data.frame(
trial = rep(1:2, each = 3),
focal = c("A", "A", "B", "D", "D", "E"),
other = c("B", "C", "C", "E", "F", "F"),
kin = c(1, 0, 1, 0, 1, 1), # first network
inverse_distance = c(0, 1, .5, .25, .1, 0) #second network
)
ILV_c <- data.frame(
id = LETTERS[1:6],
age = c(-1, -2, 0, 1, 2, 3), # continuous variables should be normalized
sex = as.factor(c("F", "M", "M", "M", "F", "F")), # Categorical ILVs must be input as factors
weight = c(0.5, .25, .3, 0, -.2, -.4)
)
data_list <- import_user_STb(
event_data = event_data,
networks = networks,
ILV_c = ILV_c,
ILVi = c("age"),
ILVs = c( "age", "sex"),
ILVm = c("weight")
)
model = generate_STb_model(data_list,
veff_params = c("lambda_0", "s", "age"),
veff_type=c("id", "trial"))If you inspect the model code above, you’ll see that it’s generated 2 different sets of varying effects for age, since age is being considered as additive rather than multiplicative: one as it’s applied to the intrinsic rate, and one as it’s applied to the social transmission rate.
Other model types: OADA and dTADA
So far we have been building continuous time of acquisition models—we
have measured when individuals experienced an event. You might be less
certain about the timing of events. If you have coarser data where you
know that individuals learned within a given time period, but are
uncertain of exactly when, you can create a discrete TADA (dTADA) model.
If you have no time information, but only the order of acquisition, you
can also create an OADA model. The data_type argument
allows you to choose between these options:
model_cTADA = generate_STb_model(data_list, data_type = "continuous_time") #default
model_dTADA = generate_STb_model(data_list, data_type = "discrete_time")
model_OADA = generate_STb_model(data_list, data_type = "order")A flow chart for deciding between models can be found in Hasenjager et al. 2021.
Edge weight uncertainty
Rather than using point estimates for edge weights, it is possible to
import posterior distributions of edge weights from generative network
models, such as those fit by the bisonr package or
the STRAND
package. The safest way to import this is to use a 3D array with
dimensions (e.g. association[draw,focal_ID,other_ID]). Each
value encodes the influence that other_ID has on focal_ID, so
association[1,1,2] represents the edge along which social
transmission can occur from individual 2 to 1,
and association[1,2,1] is vice versa. If the network is
undirected, these values should be equal. Edge weights should be on the
logit scale. Diagonal values (self-loops) are ignored when STbayes
processes the data and won’t influence the model. These can be set to
zero in the input data. To create a multi-network nbda, just provide a
list of arrays, one for each network. Alternatively, you can provide
single fits (or a list of fits) from STRAND or bisonr to
import_user_STb(), although depending on how the network
model was configured, this may have unexpected results. The imports work
as of the publication of the paper, but the safest method is to extract
and format the posterior draws yourself.
# network has 10 individuals, create mock diffusion data
event_data <- data.frame(
trial = 1,
#bison stores ids as characters, ids should match across dataframes
id = as.character(c(1:10)),
time = sample(1:101, 10, replace = FALSE),
t_end = 100
)
# mock 1000 posterior draws.
association <- array(rnorm(1000 * 10 * 10, mean = 0, sd = 1),
dim = c(1000, 10, 10))
data_list <- import_user_STb(event_data, networks = association)
model <- generate_STb_model(data_list)
fit_STb(data_list = data_list,
model_obj = model)
# it's also possible to do multi-network models
visual_contact <- array(rnorm(1000 * 10 * 10, mean = 0, sd = 1),
dim = c(1000, 10, 10))
data_list <- import_user_STb(event_data,
networks = list(association, visual_contact))
model <- generate_STb_model(data_list)Importing from bisonr:
bisonr_fit <- STbayes::bisonr_fit
# network has 10 individuals, create mock diffusion data
event_data <- data.frame(
trial = 1,
#bison stores ids as characters, ids should match across dataframes
id = as.character(c(1:10)),
time = sample(1:101, 10, replace = FALSE),
t_end = 100
)
data_list <- import_user_STb(event_data, networks = bisonr_fit)
model <- generate_STb_model(data_list)
# it's also possible to do multi-network models.
# here i add two of the same just to illustrate
data_list <- import_user_STb(event_data,
networks = list(bisonr_fit, bisonr_fit))
model <- generate_STb_model(data_list)Importing from STRAND:
strand_fit <- STbayes::strand_results_obj
# network has 10 individuals, create mock diffusion data
event_data <- data.frame(
trial = 1,
#bison stores ids as characters, ids should match across dataframes
id = as.character(c(1:10)),
time = sample(1:101, 10, replace = FALSE),
t_end = 100
)
data_list <- import_user_STb(event_data, networks = strand_fit)
model <- generate_STb_model(data_list)
# multi-network models.
data_list <- import_user_STb(event_data,
networks = list(strand_fit, strand_fit))
model <- generate_STb_model(data_list)Complex transmission
STbayes can be used to fit and create models of complex transmission.
You can create a log-likelihood that includes a frequency-dependent
transmission rules by using the transmission_func argument
of generate_STb_model:
data_list = import_user_STb(STbayes::event_data, STbayes::edge_list)
model_standard = generate_STb_model(data_list, transmission_func="standard") # default
model_f = generate_STb_model(data_list, transmission_func="freqdep_f")
model_k = generate_STb_model(data_list, transmission_func="freqdep_k")The package includes a “freqdep_f” model of frequency-dependent bias as:
STbayes also includes an alternative parameterization “freqdep_k” based on a scaled version of Dino Dini’s normalized tunable sigmoid function. In the first case, f<1 would be evidence of an anti-conformist transmission bias, f=1 would be proportional, and f>1 would be conformist. In the second case, the shape parameter k < 0 is conformist, k=0 proportional, and k>0 anti-conformist. Both parameterizations create a similar relationship between the proportion of informed neighbours and the weight of that information on the rate of an event happening for a given individual:
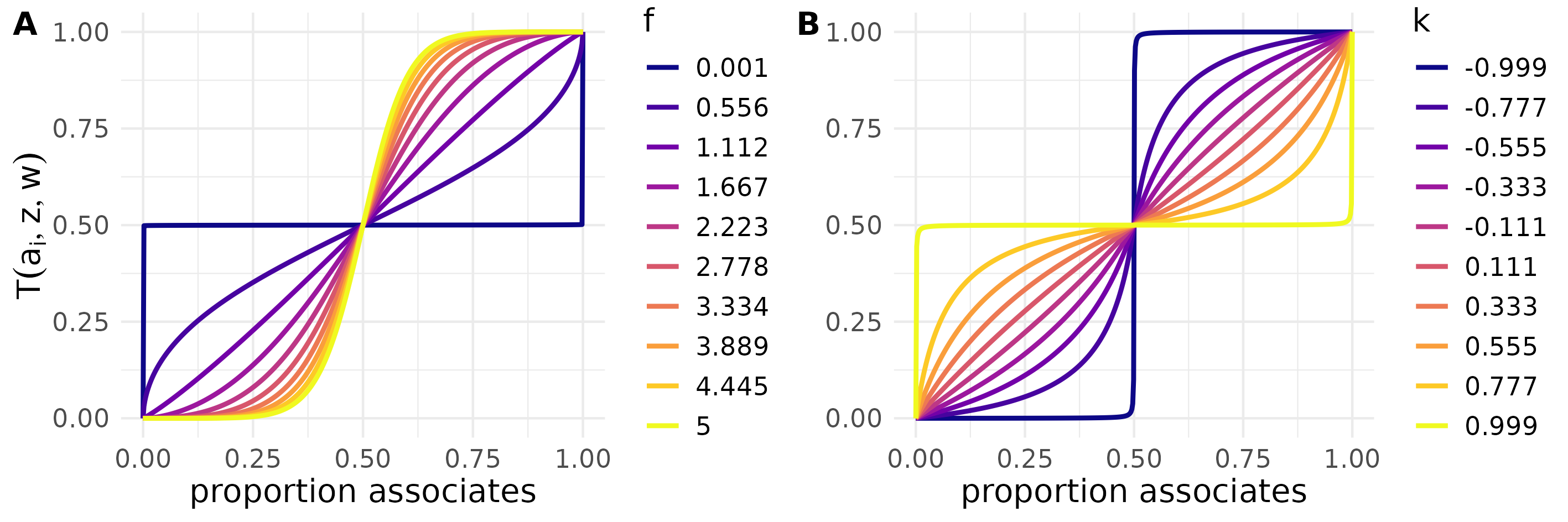
You might find that one or the other has convergence issues, so we provide both. Complex transmission can be implemented with varying effects, ilvs, oada and ctada type models, etc. We note however that the interpretation of the parameter changes when fitting a complex transmission model, as it is no longer scaled per unit connection, but rather per the proportion of associates. NB: You can set varying effects on or but tbh that’s gonna need a lot of data to estimate.
Setting priors
You might want to customize the priors of your model. Here are the default priors:
default_priors <- list(
log_lambda0 = "normal(-4, 2)", # intrinsic rate
log_sprime = "normal(-4, 2)", # social transmission rate
beta_ILV = "normal(0,1)", # beta coefficients for ILVs
log_f = "normal(0,1)", # exponent parameter under freqdep_f models
k_raw = "normal(0,3)", # k parameter under freqdep_k models
z_veff = "normal(0,1)", # varying effects
sigma_veff = "normal(0,1)", # SD of varying effects
rho_veff = "lkj_corr_cholesky(3)", # LKJ prior for veff corr matrix
gamma = "normal(0,1)" # gamma parameter for Weibull intrinsic hazard model
)To adjust any one of them, call generate_STb_model and provide a named list of those you wish to change in the priors argument. The distributions should be written with the usual Stan convention. You only need to supply those which you wish to change. For example, you might want to tighten your priors on intrinsic rate, social rate and beta params for ILVs.
data_list = import_user_STb(STbayes::event_data, STbayes::edge_list)
model <- generate_STb_model(data_list, priors = list(
log_lambda0 = "normal(-4, 1)",
log_sprime = "uniform(-4, 1)",
beta_ILV = "normal(0, .5)"
))At the moment, the function will always print all priors that are possible to specify, regardless of the type of model you are creating. At least this makes it easy to reference the names when you need them.
High-resolution data mode
If you have very high-frequency sampling of behaviour (e.g. from a motion capture system), you can take advantage of it to create a model with dynamic networks and dynamic transmission weights that can change in value frequently between events. Sampling intervals should be regular (e.g. 30 fps). Below is a dataset where networks and transmission weights are recorded every time-step, rather than summarized per inter-event interval. If fitting a model with standard transmission, STbayes preprocesses the data for efficient fitting.
data_hires <- import_user_STb(
event_data = STbayes::events_hires,
networks = STbayes::networks_hires,
t_weights = STbayes::t_weights_hires,
high_res = T
)
model_hires <- generate_STb_model(data_hires, transmission_func = "standard")If you plan to fit a model with complex transmission, it’s not
possible to pre-process the data because of the non-linearity of the
transmission rule. You should use import_user_STb2().
High-res complex transmission models will take significantly longer to
fit as they loop through each time-step, rather than inter-event
intervals.
data_hires <- import_user_STb2(
event_data = STbayes::events_hires,
networks = STbayes::networks_hires,
t_weights = STbayes::t_weights_hires,
high_res = T
)
model_hires <- generate_STb_model(data_hires, transmission_func = "freqdep_k")Import NBDA data objects
STbayes is compatible with most NBDA objects. The function will try
to figure out what was meant to be done with ILVs. However, some
features of STbayes require using import_user_STb. Below we
read in an NBDA object (taken from Tutorial 4.1 from Hasenjager et
al. 2021):
nbdaData_cTADA <- STbayes::tutorial4_1
data_list = import_NBDA_STb(nbdaData_cTADA)
str(data_list)